Z Gaibee et al. N Engl J Med 2025;392:1297-1309. The Genetic Architecture of Congenital Diarrhea and Enteropathy
Background:”Congenital diarrhea and enteropathies (CODEs) are a group of rare disorders that primarily affect the function of intestinal epithelial cells, leading to infantile-onset diarrhea and poor growth. Molecular defects in CODEs can be classified into six categories: epithelial trafficking and polarity, immune-cell-regulation, nutrient and electrolyte transport, enteroendocrine-cell development, nutrient metabolism, and other. CODEs are associated with substantial morbidity and mortality. Patients often receive lifelong fluid and nutritional management. Genetic causes include pathogenic variants in MYO5B (microvillus inclusion disease), EPCAM (tufting enteropathy), NEUROG3 (enteric anendocrinosis), DGAT1 (protein-losing enteropathy), and SLC9A3 (congenital sodium diarrhea). Treatment options are currently limited. However, an understanding of some of the genetic causes of CODEs has led to targeted therapies such as dietary treatments and the development of preclinical pharmacologic treatments.”
Methods: In this case series with 129 infants, the authors analyzed the exomes or genomes of infants with suspected monogenic congenital diarrheal disorders. Using cell and zebrafish models, we tested the effects of variants in newly implicated genes.
Key findings:
- Causal genetic variants were identified in 62 infants (48%). This included a new founder NEUROG3 variant
- Using cell and zebrafish models, the authors uncovered and functionally characterized three novel genes associated with CODEs: GRWD1, MYO1A, and MON1A
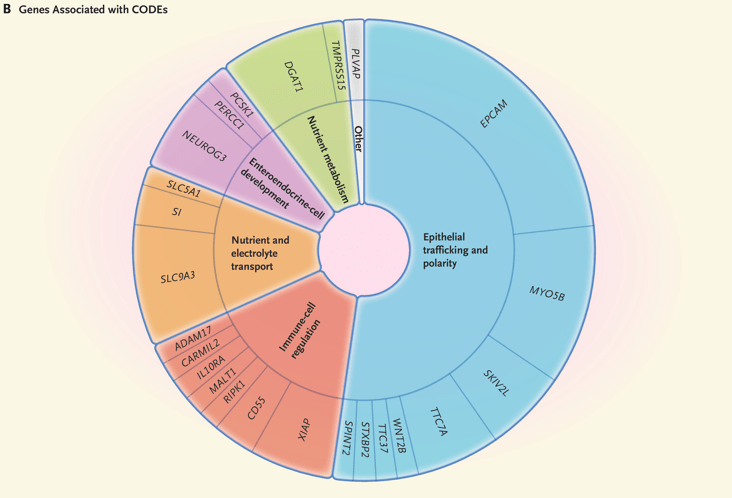
My take: Exome sequencing is an important part of the evaluation of infants with CODEs
Related blog posts:
